BOAR for TrPL fitting
Version 1.0.0 (c) Vincent M. Le Corre, Larry Lueer, i-MEET 2021-2023
This notebook is made to use BOAR for fitting transient photoluminescence (TrPL) data with rate equations.
Here we use the following rate equations:
\[\frac{dn}{dt} = G - k_{trap} n (Bulk_{tr} - n_t) - k_{direct} n (p + p_0)\]
\[\frac{dn_t}{dt}= k_{trap} n (Bulk_{tr} - n_t) - k_{detrap} n_t (p + p_0)\]
\[\frac{dp}{dt} = G - k_{detrap} n_t (p + p_0) - k_{direct} n (p + p_0)\]
where \(n\) and \(p\) are the electron and hole charge carrier densities, \(G\) is the generation rate in m⁻³ s⁻¹, ktrap and kdetrap are the trapping and detraping rates in m³ s⁻¹, and kdirect is the bimolecular/band-to-bad recombination rate in m³ s⁻¹.
[1]:
# Activate matplotlib widgets
# %matplotlib inline
# comment the next line if you are on the jupyterhub server
# %matplotlib widget
# %matplotlib notebook
# Import libraries
import sys,os
from numpy.random import default_rng
import warnings
warnings.filterwarnings('ignore') # comment this out to see warnings
# Import boar
# import one folder up
sys.path.append('../') # comment out if the Notebook is in the Notebooks folder
from boar import *
[2]:
# Define the path to the data directory
curr_dir = os.getcwd()
parent_dir = os.path.abspath(os.path.join(curr_dir, '../')) # path to the parent directory
res_dir = os.path.join(parent_dir,'temp') # path to the results directory
Define the free parameters to be optimized
[3]:
# define Fitparameters
# create fit parameters for the various processes we have to consider
True_values = {'kdirect': 1e-17, 'ktrap': 5e-18, 'kdetrap':1e-18,'Bulk_tr':1e23,'p_0':0e19, 'QE': 0.9, 'I_PL': 1e-32, 'N0': 1.04103e24}
params = []
kdirect = Fitparam(name = 'kdirect', val = True_values['kdirect'], relRange = 1, range_type = 'log',
lim_type = 'relative',optim_type='log', display_name = 'k$_2$',unit='m$^{3}$ s$^{-1}$')
params.append(kdirect)
ktrap = Fitparam(name = 'ktrap', val = True_values['ktrap'] , relRange = 1, range_type = 'log',
lim_type = 'relative',optim_type='log', display_name = 'k$_{trap}$',unit='m$^{3}$ s$^{-1}$')
params.append(ktrap)
kdetrap = Fitparam(name = 'kdetrap', val = True_values['kdetrap'], relRange = 1, range_type = 'log',
lim_type = 'relative',optim_type='log', display_name = 'k$_{detrap}$',unit='m$^{3}$ s$^{-1}$')
params.append(kdetrap)
Bulk_tr = Fitparam(name = 'Bulk_tr', val = True_values['Bulk_tr'], relRange = 1, range_type = 'log',
lim_type = 'relative',optim_type='log', display_name = 'N$_T$',unit='m$^{-3}$')
params.append(Bulk_tr)
p_0 = Fitparam(name = 'p_0', val = True_values['p_0'], relRange = 0, range_type = 'log',
lim_type = 'relative',optim_type='log', display_name = 'p$_0$',unit='m$^{-3}$')
params.append(p_0)
QE = Fitparam(name = 'QE', val = True_values['QE'], lims = [0.02, 1], relRange = 0, range_type = 'linear',
lim_type = 'absolute',optim_type = 'linear', display_name = 'QE',unit='%')
params.append(QE)
I_PL = Fitparam(name = 'I_PL', val = True_values['I_PL'], lims = [1e-33, 1e-31], relRange = 0, range_type = 'linear',
lim_type = 'absolute',optim_type = 'linear', axis_type='log',display_name= 'I$_{PL}$',unit='a.u.')
params.append(I_PL)
N0 = Fitparam(name = 'N0', val = True_values['N0'], lims = [5e21, 5e22], relRange = 0, range_type = 'log',
lim_type = 'absolute',optim_type = 'log', axis_type='log',display_name= 'N$_0$',unit='m$^{-3}$')
params.append(N0)
Prepare fake data for fitting
In the next block we create some fake data with some random noise and plot it.
[4]:
# Make fake data
rng = default_rng()
noise = 1e3 # noise level
fpu = 1e3 # pump frequency in Hz
background = 0e20 # background pump density in m^-3
# create a pump pulse
x1 = np.geomspace(1e-8, 50e-6, num=10000)
#add 0 to the beginning of the pump pulse
x1 = np.insert(x1,0,0)
X_dimensions = ['t','Gfrac']
X,y,weight = [],[],[]
trPL = TrPL_agent()
trPL.trPL_model = partial(Bimolecular_Trapping_Detrapping_equation, solver_func='odeint', method='Radau')
trPL.pump_params['fpu'] = fpu
trPL.pump_params['background'] = background
Gfracs = np.geomspace(1e-3,1,endpoint=True,num=5)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10,10))
for Gfrac in Gfracs:
X1 = [[xx, Gfrac] for xx in x1]
X = X + X1
y1 = trPL.trPL(np.asarray(X1), params, X_dimensions=X_dimensions,take_log=True)
y1 = y1
y = y + list(y1)
weight = weight + list(1/(np.abs(max(y1)*np.ones(len(y1))))) #
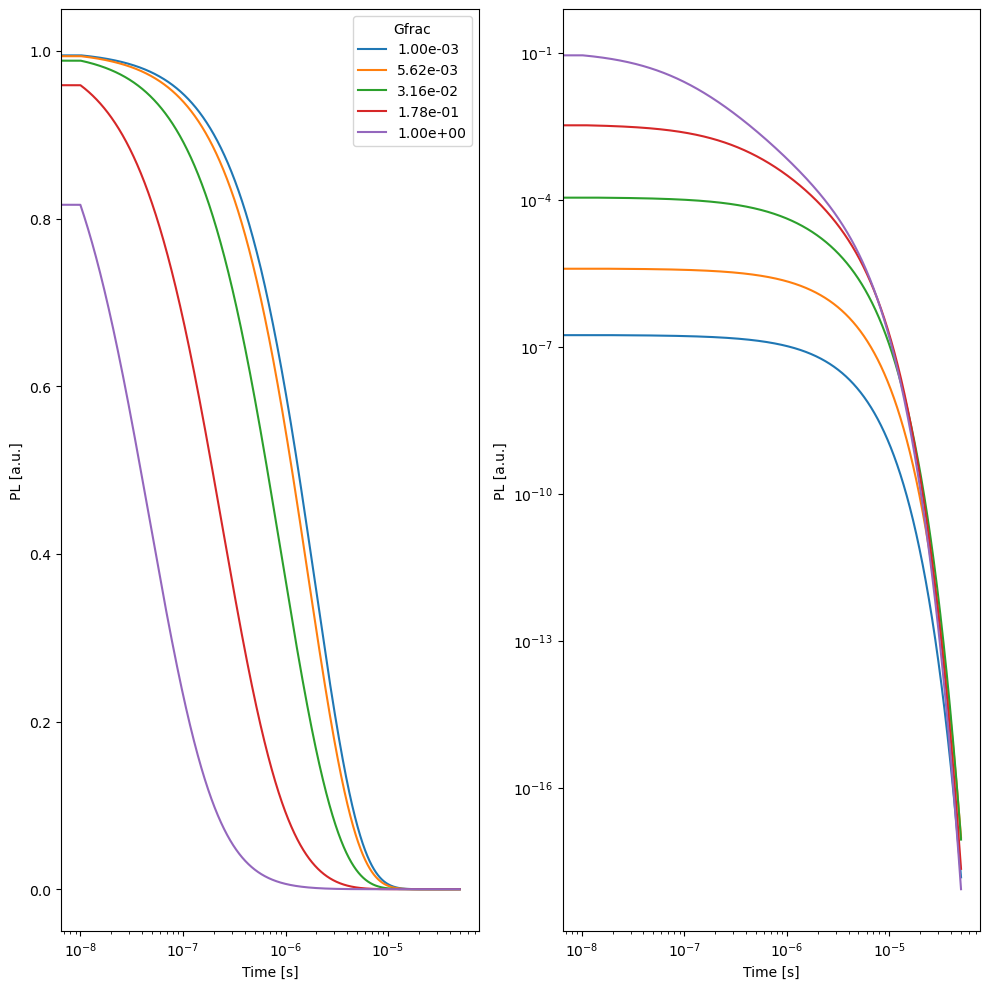
ax1.semilogx(x1,10**y1/max(10**y1),label=f'{Gfrac:.2e}')
ax2.loglog(x1,10**y1)
ax1.set_ylabel('PL [a.u.]')
ax1.set_xlabel('Time [s]')
ax1.legend(title='Gfrac')
ax2.set_ylabel('PL [a.u.]')
ax2.set_xlabel('Time [s]')
plt.tight_layout()
X = np.array(X)
y = np.array(y)
weight = np.array(weight)
weight/=np.mean(weight)
weight[weight>10]=10
Start the optimization
[5]:
# start multiobjective optimization
X_dimensions = ['t','Gfrac']
y_dimension = 'PL [a.u.]'
target = {'model':partial(trPL.trPL,X_dimensions=X_dimensions,take_log=True), 'target_name':'trPL',
'data':{'X':X,'y':y,'X_dimensions':X_dimensions,'X_units':['s',''],'y_dimension':y_dimension,'y_unit':''}
,'weight':weight,'target_weight':1}
targets = [target]
mo = MultiObjectiveOptimizer(targets = targets, params=params, res_dir=res_dir)
# define the optimization parameters
n_jobs = 4
n_jobs_init = 20
n_yscale= 20
n_initial_points = 80
n_BO = 150
n_BO_warmstart = 60
mo.warmstart = 'None' # whether to use warmstart to recall or collect points or not
mo.SaveOldXY2file = os.path.join(res_dir,'old_XY.json') # path to the file where old points are saved
mo.Path2OldXY = os.path.join(res_dir,'old_XY.json') # path to the file where old points are saved
kwargs = {'check_improvement':None,'max_loop_no_improvement':10,'xtol':1e-3,'ftol':1e-3}
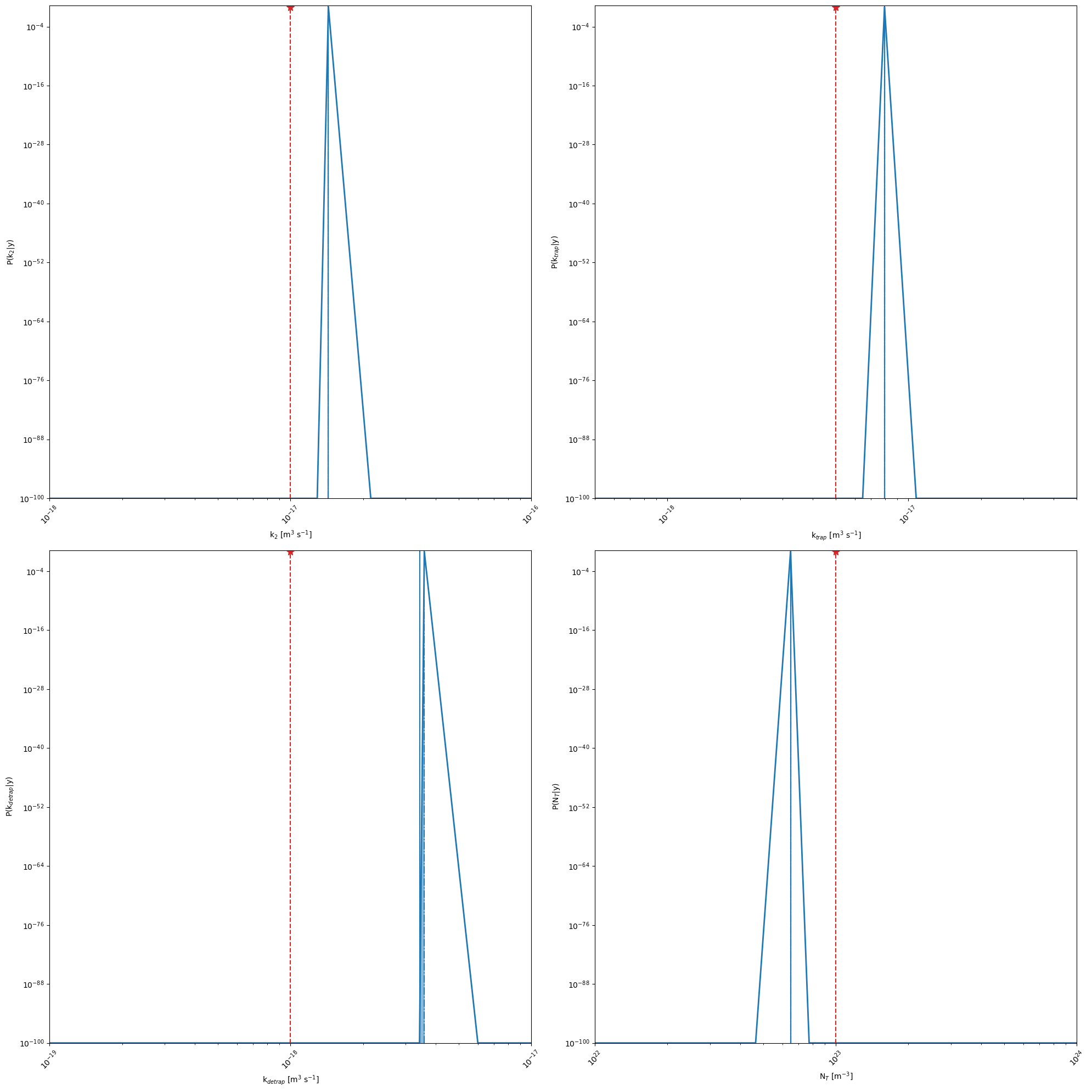
kwargs_posterior = {'Nres':10,'Ninteg':1e5,'logscale':True,'vmin':1e-100,'zoom':0,'min_prob':1e-40,'clear_axis':False,'show_points':True,'savefig':True,'figname':'param_posterior','True_values':True_values}
kwargs_plot_obj = {'zscale':'linear'}
r = mo.optimize_sko_parallel(n_jobs=n_jobs,n_yscale=n_yscale, n_BO=n_BO, n_initial_points = n_initial_points,n_BO_warmstart=n_BO_warmstart,n_jobs_init=n_jobs_init,kwargs=kwargs,verbose=False,loss='linear',threshold=1000,base_estimator = 'GP',show_objective_func=False,show_posterior=True,kwargs_posterior = kwargs_posterior,kwargs_plot_obj=kwargs_plot_obj)
Starting with initial points
Initial points done in 6.65 s
Starting with BO
BO done in 467.19 s
Ground truth minimum at: [-16.842545996331125, -17.09920231621749, -17.464237847358547, 22.811653035750375] with function value: 0.008576118354324761
Minimum of surrogate function: [-16.83767283643464, -17.097937121339612, -17.379615854523234, 22.81424355001567] with function value 0.007834800280307697
[6]:
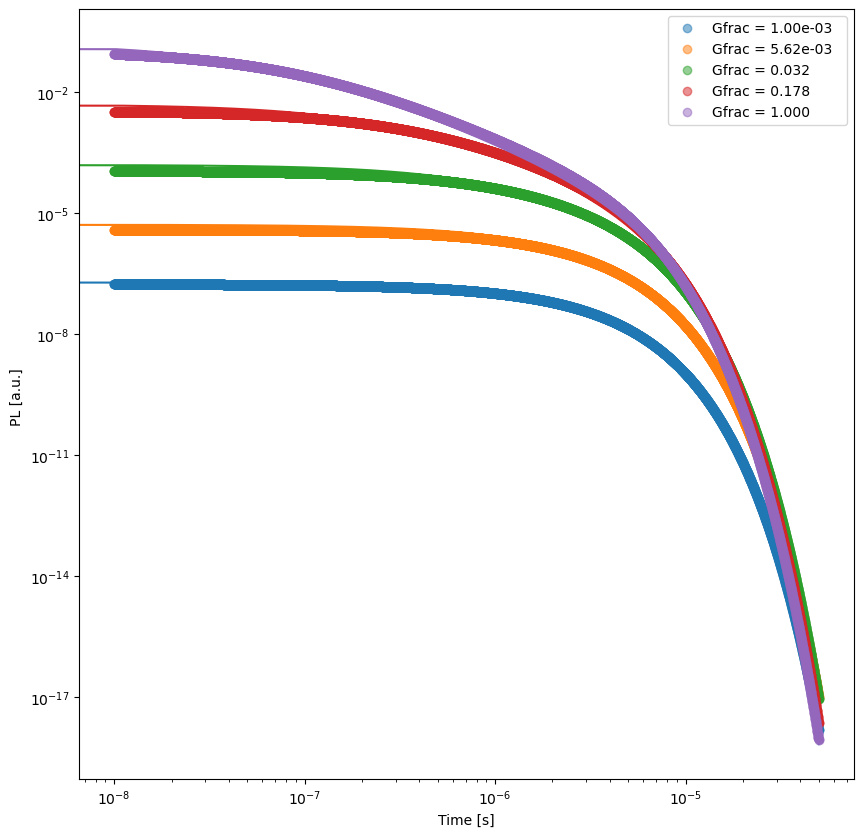
# Plot and save the results
fit_results = []
kwargs_plot_res = {'x_scaling':1,'xaxis_label':'Time [s]','xscale_type':'log','yscale_type':'log','norm_data':False,'delog':True,'figsize':(10,10)}
for num,t in enumerate(targets):
kwargs_plot_res['figname'] = os.path.join(res_dir,t['target_name']+f'_fit_{num}')
trPL.plot_fit_res(t,mo.params,'t',xlim=[],ylim=[],kwargs=kwargs_plot_res)
X = t['data']['X']
y = t['data']['y']
X_dimensions = t['data']['X_dimensions']
yfit = t['model'](X,mo.params,X_dimensions=X_dimensions) # get the best fits
data = np.concatenate((X, y.reshape(len(y),1), yfit.reshape(len(yfit),1)), axis=1)
fit_results.append(data)
# prepare the data for saving
param_dict = trPL.get_param_dict(mo.params) # get fitparameters (and fixed ones)
pout = [[f'{v:.3E}' if isinstance(v,float) else v for _,v in pp.items()] for pp in param_dict]
save_output = False
if save_output:
# produce output excel file with data, fitparameters and FOMs
fn_xlsx = 'fits_results.xlsx'
namecols = X_dimensions + ['Jexp','Jfit']
# delete old file if it exists
if os.path.exists(os.path.join(res_dir,fn_xlsx)):
os.remove(os.path.join(res_dir,fn_xlsx))
with pd.ExcelWriter(os.path.join(res_dir,fn_xlsx), mode='w') as writer:
for i,t in enumerate(targets):
if 'target_name' in t.keys():
tname = t['target_name']
else:
tname = 'data'
namecols = X_dimensions + [tname+'_exp',tname+'_fit']
df = pd.DataFrame(fit_results[i],columns=namecols)
df.to_excel(writer, sheet_name = tname+f'_{i}')
df = pd.DataFrame(pout,columns=[k for k in param_dict[0].keys()])
df.to_excel(writer, sheet_name = f'params')
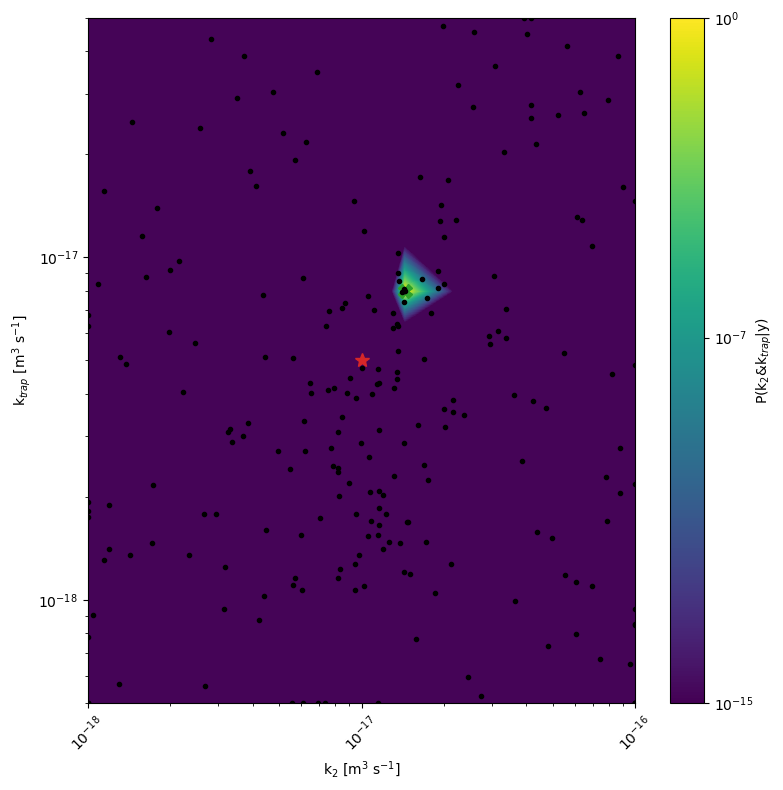
[7]:
mo.marginal_posterior_2D('kdirect', 'ktrap', Nres=10, Ninteg=1e5, logscale=True,points=mo.points, True_values=True_values,vmin=1e-15)
[8]:
# Clean output files from simulation folders
from boar.SIMsalabim_utils.CleanFolder import *
Do_Cleaning = False # Careful, this will delete all files in the folder
if Do_Cleaning:
# delete old_xy.json file if it exists
# os.remove(mo.path2oldxy) # remove the old_xy.json file if it exists
# delete warmstart folder if it exists
if os.path.exists(os.path.join(os.getcwd(),'warmstart/')):
shutil.rmtree(os.path.join(os.getcwd(),'warmstart/'))
# delete temp folder if it exists
if os.path.exists(os.path.join(os.getcwd(),'temp/')):
shutil.rmtree(os.path.join(os.getcwd(),'temp/'))